Overview of the kinase family
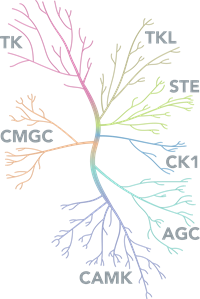
Kinases Diagram
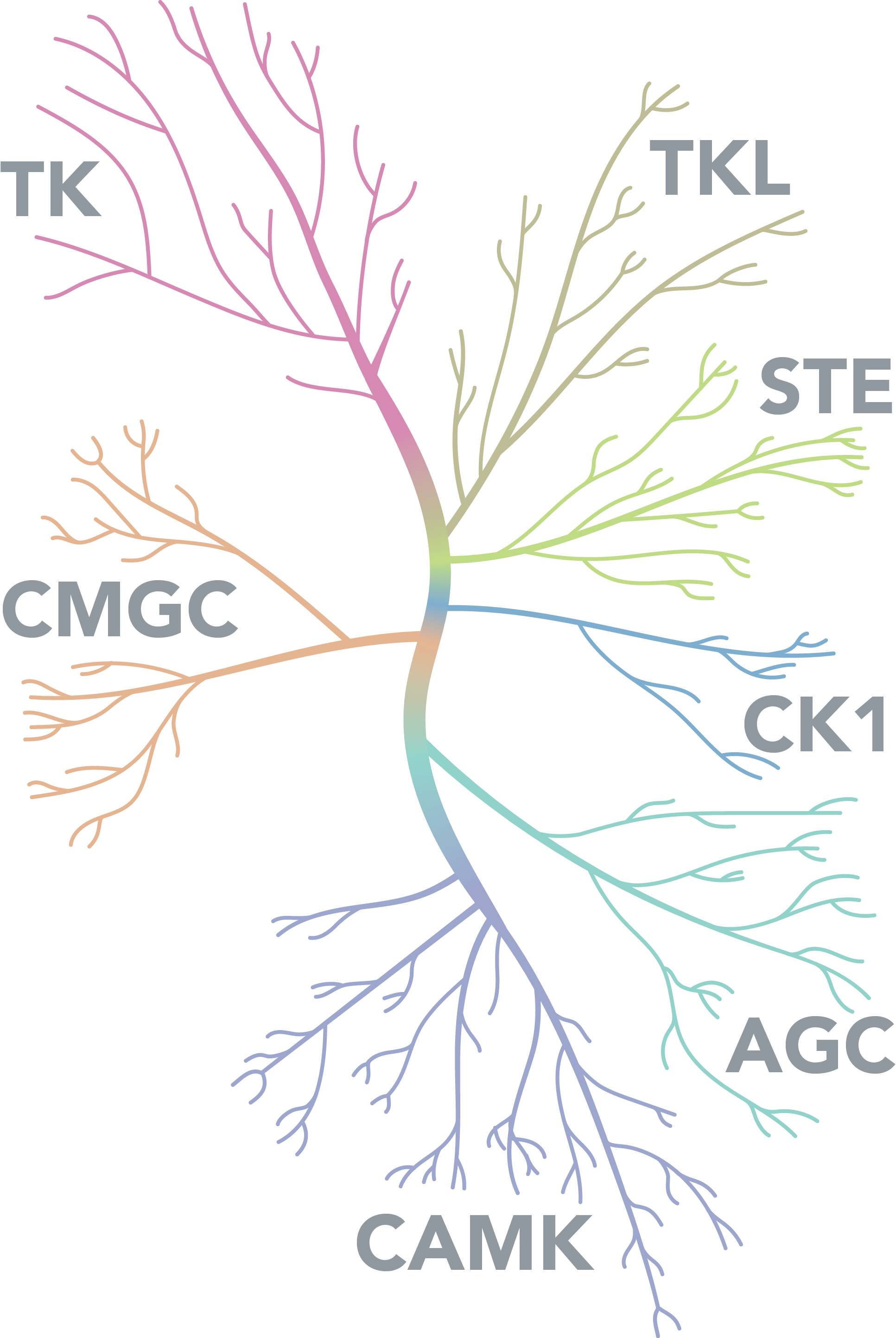
Kinases are a large family of enzymes
Kinases Diagram
Kinases are a large family of enzymes with over 500 encoded in the human genome and all share a highly conserved kinase domain. Dependent on the substrate they act upon they can be broadly classified into 3 categories: protein kinases, carbohydrate kinases and lipid kinases. Due to the diverse roles that kinases play in the cell they are implicated in a number of diseases including cancer:
Protein kinases
- can phosphorylate substrate proteins on serine, threonine, tyrosines or histidine residues. Mitogen-activated protein kinases such as RAF kinases involved in the MAPK signaling pathways and cyclin dependent kinases (CDKs) involved in cell cycle regulation are prime examples of pathways implicated in cancers.
Lipid kinases
- alter the localization and reactivity of lipids through phosphorylation. A well-known example of a lipid kinase is phosphoinositide 3-kinase which is implicated in a number of cancers, mainly through alterations to its catalytic subunit - PIK3CA.
Carbohydrate kinases
Order products
Human knockout HAP-1 cells
The single largest bank of isogenic cell lines with over 7,500 cell lines to choose from and trusted by academia, biotech, and pharma research labs.
Cancer-related cell lines
Choose from over 300 knock-in and knockout cell line models in many standard cancer cell lines such as DLD1, MCF10A, and HCT116.
Cas9 Stable Cell Lines
Simplify gene editing experiments with stably expressing Cas9 cell lines
CRISPRmod CRISPRa dCas9-VPR Stable Cell Lines
Streamline CRISPR activation experiments with stably expressing dCas9-VPR cell lines
Helpful Resources
Cell Line Engineering Brochure
Save time and de-risk your project. With 7,500 readymade knockouts from Horizon you can validate your research without having to invest valuable time, money, and resources.
Learn more
Top peer reviewed scientific articles using HAP1 cell lines
Need help?
Can't find the right cell model for your research?
Our Express and Custom engineering services could help you.
