The Dharmacon™ CRISPRmod platform of gene modulation tools promote (CRISPRa) or repress (CRISPRi) target gene transcription, without cutting the DNA.
What is CRISPRmod?
CRISPRmod is the Dharmacon family of CRISPR activation and CRISPR interference reagents, used for targeted modulation of endogenous gene expression.
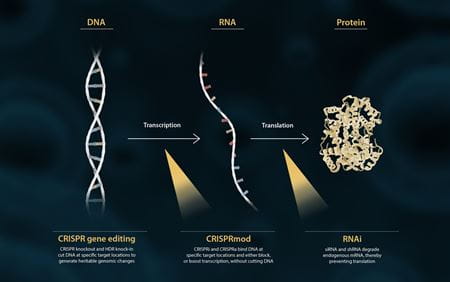
In the central dogma of molecular biology, DNA is transcribed into RNA, which is then translated into protein (figure 1). Researchers can modulate this pathway at multiple points through loss- or gain-of-function experiments to better understand gene function.
Existing CRISPR knockout and HDR knock-in techniques allow researchers to edit (make heritable changes to) cellular DNA, while traditional RNAi techniques degrade mRNA, thereby preventing translation.
CRISPRmod offers a fresh gene modulation method, regulating expression at the transcriptional level. The Dharmacon CRISPRmod portfolio includes both CRISPRa and CRISPRi systems, to respectively up- or down-regulate target gene expression.
How does CRISPRmod work?
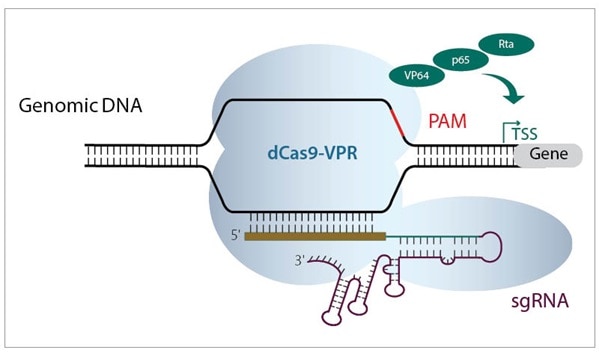
Both CRISPRmod systems (CRISPRa and CRISPRi) use a deactivated Cas9 construct fused to either activators or repressors, to leverage the power of PAM-anchored targeting for transcriptional modulation (figure 2). CRISPRmod simply brings the precision of CRISPR to gene modulation.
The native Cas9 DNA-cutting functionality has been obliterated by point mutations in the RuvCI and HNH nuclease domains (W. Cheng et al.), changing the protein to a deactivated, or dead, Cas9 (dCas9). This dCas protein is further engineered for targeted transcriptional modulation through the fusing of activator or repressor domains to the C-terminus.
CRISPRa system |
CRISPRi system |
 |
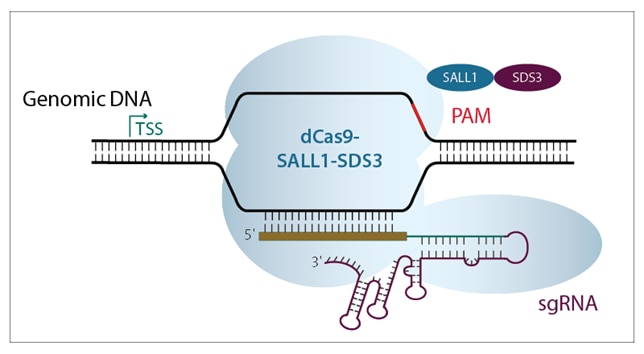 |
Figure 2. Schematic diagrams showing a) CRISPRa machinery binding upstream of TSS to activate transcription, and b) CRISPRi machinery binding downstream of target gene TSS to block transcription.
Why use CRISPRmod?
CRISPRmod enables a new generation of CRISPR-based experiments for endogenous gene modulation. CRISPRmod also offers an orthogonal method for confirming phenotypes observed from other gene modulation methods.
- Activate or inhibit genes within their native context for more biologically relevant models
- Simultaneously modulate multiple genes to better understand a pathway or network
- Orthogonally validate CRISPR knockout or siRNA results for more robust datasets
Multiplexed gene modulation
CRISPRmod offers a unique method for the simultaneous modulation of multiple genes. Each guide RNA binds to its specific DNA target independently and relies on an exogenously supplied dCas9 fusion protein, thereby minimizing any competition for endogenous pathways. Here we show the effectiveness of multiplexing CRISPRi within iPSC cells (figure 3).
Synthetic sgRNAs enable easy multiplexing for the simultaneous repression of multiple genes
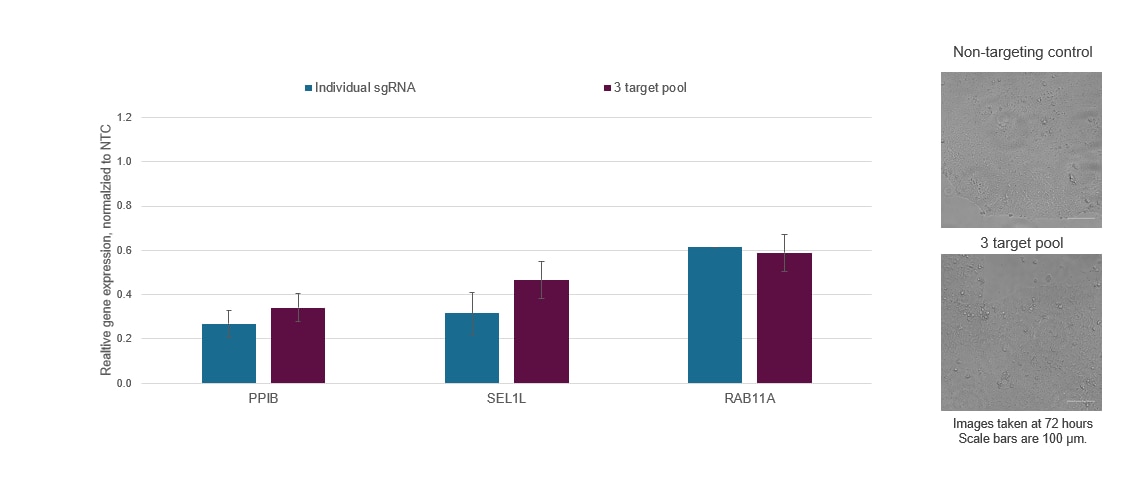
Figure 3. Individual CRISPRi sgRNAs can be pooled together in a single reagent to achieve enhanced target gene repression or multiplexed to enable the simultaneous repression of multiple genes. WTC-11 human iPS cells stably expressing integrated dCas9-SALL1-SDS3 were nucleofected with synthetic sgRNA targeting PPIB, SEL1L, and RAB11A via a Lonza 96-well Shuttle system. The most active pre-designed sgRNA for each gene target was selected and used either individually or as part of a multi-target pool at a concentration of 3 µM per guide. Cells were harvested 72 hours post-nucleofection, total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative expression for each target gene was calculated with the ∆∆Cq method using ACTB as the housekeeping gene and normalized to a non-targeting control (NTC). Three genes were simultaneously repressed without a substantial decrease in target gene repression or marked changes in cell viability and morphology.
Orthogonal validation
Validated science is good science. Dharmacon Reagents include a wide range of tools that interrogate cellular pathways, allowing inactivation of a gene at the DNA level, modulating RNA transcription, or by degrading mature mRNA transcripts. We are here to help you choose the best method of gene activation or inactivation for your application. Better yet, choose two methods for robust, elegant, validated science.
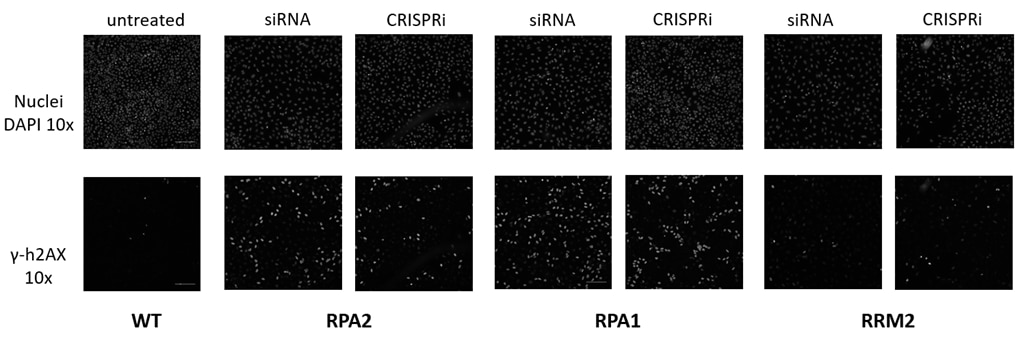
CRISPRmod presents an ideal opportunity for follow-on studies after RNAi or CRISPRko screening (figure 4). Here we used CRISPRi to follow up on an arrayed siRNA screen we had previously conducted.
Synthetic CRISPRi reagents can be used to orthogonally validate hits from siRNA screens
Figure 4. Damage Response Assay. h4AX (a constituent of core histone complexes) surrounding double-strand breaks is rapidly phosphorylated, producing a set of responses through the DNA damage signaling network. Knockdown of proteins critical to the DNA damage pathway results in an accumulation of unrepaired double-stranded DNA breaks, and an increase in phosphorylated h4AX (γ-h4AX). Hits from a prior RNAi screen were orthogonally validated using CRISPRi reagents. U2OS cells constitutively expressing dCas9-SALL1-SDS3 under the hEF1α promoter were transfected with pooled sgRNAs (50nM) or ON-TARGETplus SMARTpools (50nM) targeting RPA2, RPA1, or RRM2 using DharmaFECT 4 Transfection Reagent. 72 hours post-transfection, the cells were fixed and stained with an anti-phospho-h4AX antibody, and Hoechst stain was used to identify nuclei. A duplicate plate was harvested, total RNA was isolated, and relative gene expression was measured using RT-qPCR. The relative expression for each target gene was calculated with the ∆∆Cq method using GAPDH as the housekeeping gene and normalized to non-targeting control (NTC).
CRISPRmod product platform
We offer CRISPRmod reagent systems for both transcriptional repression (CRISPRi) and activation (CRISPRa). This incudes a full suite of synthetic guides and mRNA options for easy electroporation/transfection and rapid results, as well as lentiviral options for difficult-to-transfect cells or extended time course experiments. See a product page below or contact our expert scientific support team to find the best product format for your needs.
CRISPRi |
CRISPRa |
|
|---|---|---|
Use |
Transcriptional repression of endogenous gene expression | Transcriptional activation of endogenous gene expression |
dCas9 protein |
dCas9-SALL1-SDS3 | dCas9-VPR |
Guide |
CRISPRi guide RNA | CRISPRa guide RNA |
| CRISPRi products | CRISPRa products |
Order Products
CRISPR interference reagents
There are many options for CRISPRi experimental conditions, explore reagents
CRISPR activation reagents
There are many options for CRISPRa experimental conditions, explore reagents.
CRISPRmod controls
CRISPRi controls for loss-of-function experiments
Helpful Resources
CRISPRmod Reading List
Publications using Horizon CRISPRa or CRISPRi
What is CRISPRa and CRISPRi?
CRISPR-Cas9 for gene overexpression and down-regulation