Horizon's CRISPRmod CRISPR activation (CRISPRa) system enables gain-of-function experiments while avoiding the use of exogenous over-expression plasmids.
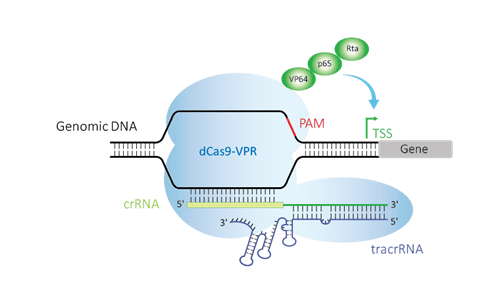
Both CRISPRmod systems (for CRISPRa and CRISPRi) use deactivated or “dead” Cas9 (dCas9), a variant of the Cas9 protein that is nuclease-deficient due to mutations in the RuvCI and HNH domains1. The CRISPRa system utilizes dCas9-VPR where the dCas9 is fused to three transcriptional activators VP64, p65, and Rta (VPR) on the C-terminus2.
Guide RNAs targeted upstream of the gene's transcriptional start site (TSS) can bind the dCas9-VPR and guide the complex to the DNA target site and enable up-regulation of the target gene. CRISPRa guide RNA designs are based on a published CRISPRa v2 algorithm3 and are different than the Edit-R guide RNAs which are optimized for functional gene knockout. The CRISPRa guide RNA can be either single guide RNA (sgRNA) or synthetic CRISPR RNA (crRNA) complexed with a trans-activating CRISPR RNA (tracrRNA) (Figure 1) enabling development of wide range of phenotypic assays for functional gene analysis.
Figure 1. CRISPRa uses deactivated Cas9 (dCas9) fused to the transcriptional activators VP64, p65, and Rta (VPR), guided by crRNA:tracrRNA upstream of a transcriptional start site (TSS) to up-regulate expression of a target gene.
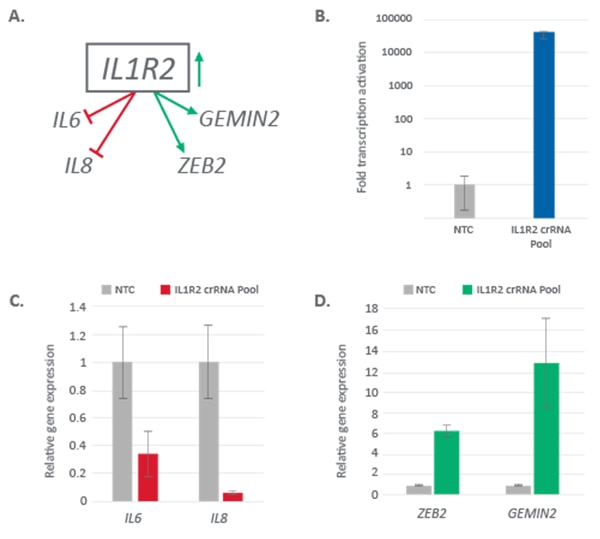
Analysis of downstream gene expression following IL1R2 activation
IL1R2 gene encodes for a cytokine receptor that belongs to the interleukin 1 receptor family. IL1R2 is a negative regulator of interleukin 1 (IL1) signaling in several ways (reviewed in4). It is competing with interleukin 1 receptor, type I (IL1R1) for IL1 ligand and it complexes with interleukin 1 receptor accessory protein (IL1RAP), thereby sequestering both the ligand and the accessory protein required for signal transduction that leads to production of cytokines IL6 and IL85, 6, 7. Other studies have shown that IL1R2 plays role in regulation of cell morphology and migration and some of its functions are exuded through regulation of ZEB2 and GEMIN2 expression levels8, 9 (Figure 2A).
In our study, we looked at downstream effects of IL1R2 transcriptional activation by CRISPRa in U2OS cells that stably express dCas9-VPR. IL1R2 is not expressed in U2OS cells, and upon transfection with an CRISPRa crRNA pool (four individual synthetic crRNAs) targeting the promoter region we observe strong activation of more than 10,000 fold (Figure 2B). This IL1R2 up regulation leads to down-regulation of IL6 and IL8 (Figure 2C) and up-regulation of ZEB2and GEMIN2 (Figure 2D), confirming literature predicted downstream gene effects and demonstrating that signaling of IL1R2 can be studied using CRISPRa.
Activation of IL1R2 with CRISPRa synthetic crRNA affects expression of downstream genes
Figure 2. A. Diagram showing predicted gene expression changes upon activation of IL1R2 from previous publications. B. U2OS cells stably expressing integrated dCas9-VPR were transfected with a synthetic crRNA pool complexed with tracrRNA (25 nM) targeting IL1R2 using DharmaFECT 4 transfection reagent. Gene activation of IL1R2 was assessed at 72 hours post transfection by RT-qPCR and is shown as fold transcriptional activation compared to non-targeting control (NTC) C. Relative expression of IL6 and IL8 upon CRISPRa of IL1R2 analyzed by RT-qPCR and normalized to NTC samples D. Relative expression of ZEB2 and GEMIN2 upon CRISPRa activation IL1R2 analyzed by RT-qPCR and normalized to NTC samples.
Highlights
- CRISPRa enables researchers to up-regulate any gene in its endogenous context, even from non-detectable levels
- CRISPRa synthetic crRNAs cause strong target gene activation and can be used for functional gene analysis suitable for short-term assay development in an arrayed format
- CRISPRa mediated gene activation leads to up- or down-regulation of other downstream genes in the signaling pathway
Authors: Eldon Chou, Maren Mayer Gross and Žaklina Strezoska
Get started now with CRISPRa »
References
- M. Jinek et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 377, 816-21 (2012).
- A. Chavez et al., Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 12, 326-8 (2015).
- M. A. Horlbeck et al., Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016).
- V. A. Peters et al., IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun. 32, 1–8 (2013).
- D. Malinowski et al., Interleukin-1 receptor accessory protein interacts with the type II interleukin-1 receptor. FEBS Lett. 3, 299-302 (1998).
- D. Lang et al., The type II IL-1 receptor interacts with the IL-1 receptor accessory protein: a novel mechanism of regulation of IL-1 responsiveness. J Immunol. 12, 6871-7 (1998).
- J. E. Sims et al., Interleukin 1 signaling occurs exclusively via the type I receptor. P Natl Acad Sci USA. 90, 6155-9 (1993).
- O. Leshem et al., TMPRSS2/ERG Promotes Epithelial to Mesenchymal Transition through the ZEB1/ZEB2 Axis in a Prostate Cancer Model. PLoS ONE. 6, e21650 (2011).
- S. Y. Chang et al., Ectopic expression of interleukin-1 receptor type II enhances cell migration through activation of the pre-interleukin 1alpha pathway. Cytokine. 45, 32-8 (2009).
Additional resources
-
What is CRISPRa or CRISPR activation?
Get an overview of this powerful gain-of-function technology!
-
CRISPRa - Resources
Find product guides, FAQs and more.`